MycotoxinDB Implementation
The MycotoxinDB was implemented using PostgreSQL database engine, while all the scripts were written in Python. JSME were used for input molecular structure.
Submit Query

To satisfy the requirement of scientists being able to search for a specific molecular fragment or retrieve a class of risk components with structural similarities, the MycotoxinDB offers multiple retrieval methods, including full structure, fragment, similarity, maximum common substructure (MCS), and text retrieval options.
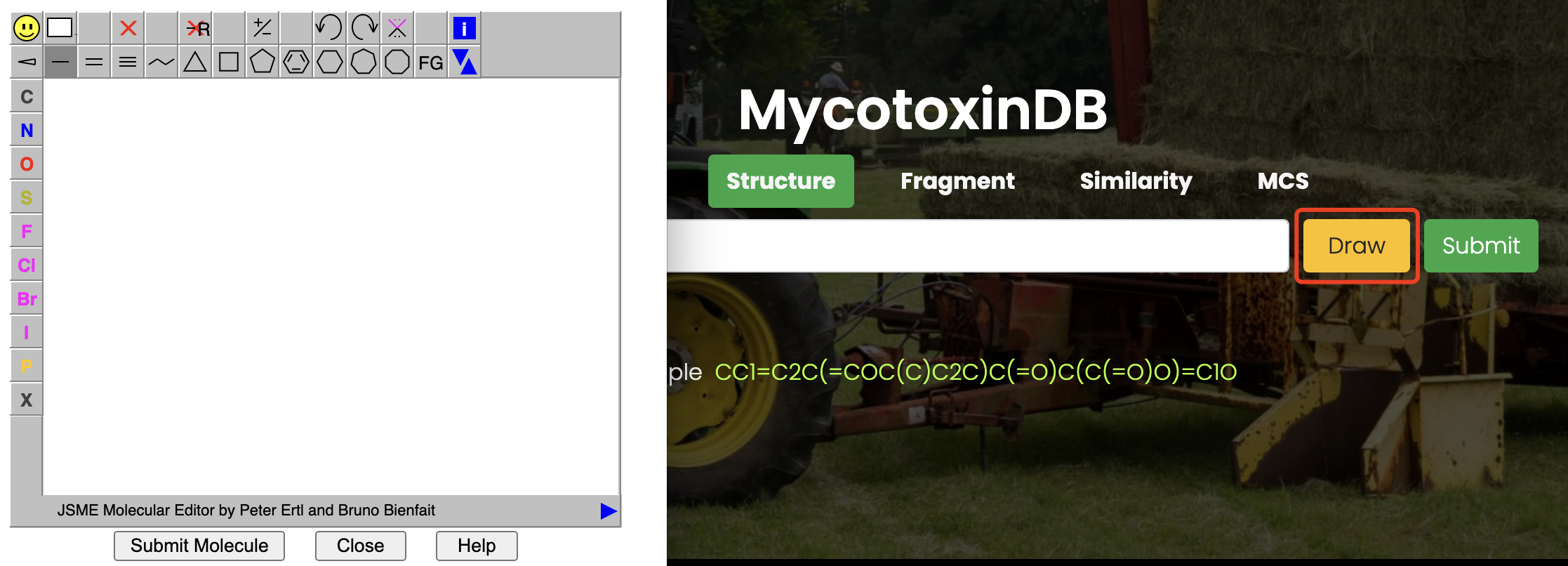
The JSME structure editor was included in the search options to enable manually editing of toxin molecules.
FAQs
Full structure retrieval means that a structural segment entered into the search by the user is fully consistent with the retrieved hit segment. To accelerate this process, we had previously normalized the molecular structure of all molecules in the MycotoxinDB. The molecular structure to be retrieved was normalized in real time and then matched to all normalized molecular structures in the MycotoxinDB, and molecules matching the exact structure were retrieved. Fragment retrieval (substructure retrieval) means that the submitted structure is a fragment and is therefore a connected subgraph of the hit structure. The similarity retrieval function employs a Tanimoto coefficient-based similarity algorithm. The retrieved molecular structure was iteratively compared to the molecules stored in the database, and the search eventually returned the 20 most similar mycotoxins. The MCS retrieval function was based on the fMCS algorithm . This retrieval method differs from similarity retrieval in that it focuses more on the complete matching of partial structural fragments rather than the similarity of the overall structure. A text retrieval method is also available for searches based on the name, CAS number, InChI, InChI key, IUPAC name, or synonyms. The MycotoxinDB includes >10,000 synonyms of mycotoxins to ensure precise search retrieval.
MycotoxinDB is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, for commercial purposes requires explicit permission of the authors and explicit acknowledgment of the source material (MycotoxinDB) and the original publication. We ask that users who download significant portions of the database cite the MycotoxinDB paper in any resulting publications.
Please send an error report to zhangdachuan@picb.ac.cnand we will verify and correct it as soon as possible. Please attach the references for the correct information if it is possible. That would be very helpful!
We recommend using Google Chrome. You can also use the latest version of other browsers.
If you have any further questions, please feel free to contact us with email jijian@jiangnan.edu.cn or zhangdachaun@picb.ac.cn .